Multisite multiomic analysis reveals diverse resistance mechanisms in end-stage ovarian cancer
An international team led by researchers from MacCallum Cancer Center in Melbourne, Australia, examined 391 tumor tissues from about 270 multisite autopsy samples collected from 15 individuals with end-stage high-grade serous ovarian cancer (HGSC). All of the provided autopsy samples came from patients who had initially responded to their cancer treatment therapies but had become resistant to them in relapse.
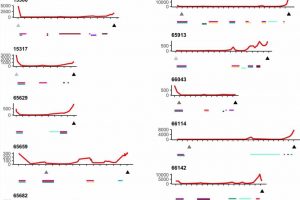
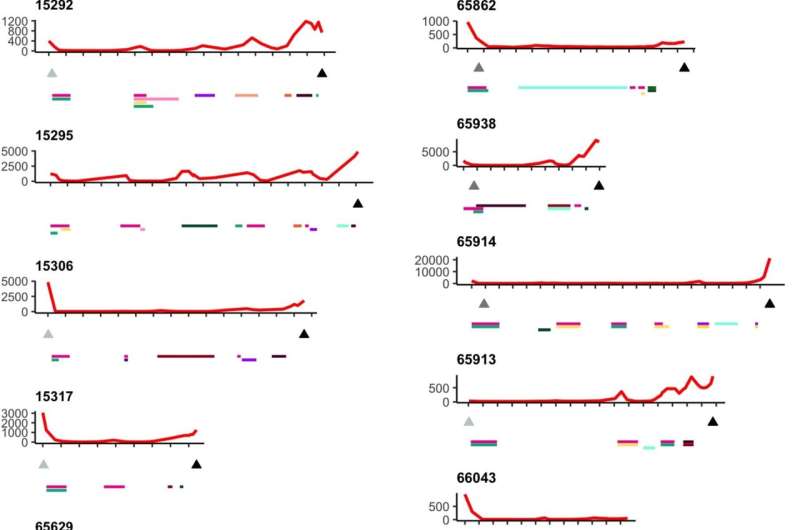
The analysis used whole-genome sequencing, targeted gene sequencing, RNA-seq, proteomics, methylation and multicolor immunofluorescence to various degrees across the collected sample. Researchers looked for variation within and between metastatic sites, focusing on the number of resistance mechanisms occurring within individual patients, and found high levels of variability, from the number of distinct clone cells and subclonal mutations to the distribution of resistance mechanisms.
All patients were found to have polyclonal disease, averaging five clones, ranging from two to nine. While cancer may begin as monoclonal, one bad cell replicating clones of itself, having many (poly) clones could mean multiple starting points of cancer from different cells. Distinct starting points can make assessing an effective therapeutic path more difficult because the differentiated clones could develop different treatment resistances. In contrast, the presence of a single clone would imply the evolution of a highly effective, singular resistance mechanism that might be able to be identified and overcome.
Targeted sequencing of resistance-related genes from spatially distinct sites within patients showed few shared mutations, and the mechanisms of resistance researchers detected were seldom present in all samples within an individual. They observed examples of convergent resistance and mechanisms of resistance that were typically subclonal. Extensive genomic variability was evident in the clones and copy number aberrations, which delete or amplify large contiguous code segments.
All samples were found to carry a somatic pathogenic TP53 gene mutation. TP53 is the gene responsible for making the protein p53, also known as the “guardian of the genome” for its ability to regulate cell division. When healthy (non-pathogenically mutated), p53 keeps cells from proliferating too quickly or chaotically. This guardian also plays a critical role in determining whether damaged DNA should be repaired or if the cell containing the damaged DNA should undergo apoptosis and self-destruct.
If the DNA is deemed fixable, p53 activates other genes to fix the damage, genes like BRCA1, BRC2, and BRIP1. All of the patients sampled in this study also had mutations in BRCA1, BRC2, and BRIP1, making them especially predisposed to cell repair errors that could lead to cancer and susceptible to multiple initial starting points for the disease.
The study indicates that multiple subclones with a diversity of mechanisms for drug resistance and immune system evasion, both varied and convergent, can be seen in end-stage disease rather than a single resistant clone. The researchers state that multisite sampling increases the power to detect novel resistance events, particularly as the selective pressure of therapy exacerbates the likelihood of unbalanced assessment from a single site.
“The challenge to therapeutic intervention in HGSC,” the researchers conclude, “cannot be overstated and reiterates the strong impetus for the development of effective treatments before acquisition of such diversity.”
The study is published in the journal Nature Genetics.
More information:
Nikki L. Burdett et al, Multiomic analysis of homologous recombination-deficient end-stage high-grade serous ovarian cancer, Nature Genetics (2023). DOI: 10.1038/s41588-023-01320-2
Journal information:
Nature Genetics
Source: Read Full Article