Study explains how the B.1.1.7 SARS-CoV-2 variant is more transmissible

The B.1.1.7 severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant was initially found in England last fall and has quickly overtaken to become the dominant coronavirus strain in the United Kingdom (UK).
The variant’s ability to increase transmission and severe infections has proven to be a challenge for public health officials looking to limit the spread.
New research led by Thushan I. de Silva from the University of Sheffield proposes that increases in subgenomic RNA (sgRNA) expression cause increased viral transmissibility and subsequent infection. They suggest the ORF9 protein, found in high amounts of sgRNA, could be regulating interferon responses.
Identifying B.1.1.7’s mechanism of action for increased transmission could help with developing or updating coronavirus treatments.
The study “Altered Sub-Genomic RNA Expression in SARS-CoV-2 B.1.1.7 Infections” is available as a preprint on the bioRxiv* server, while the article undergoes peer review.
How they did it
The researchers used the periscope tool and ARTIC Network Oxford Nanopore genomic sequencing data to analyze 4,400 positive SARS-CoV-2 nose-throat samples from healthcare workers and patients in hospitals. They also sequenced a portion of SARS-CoV-2 positive samples from the local community.
Their analysis on the B.1.1.7 variant was compared to the previously dominant variant from Spain called B.1.177.
They hypothesized that the B.1.1.7 variant had increased sgRNA expression, causing more significant phenotypic changes than other SARS-CoV-2 strains.
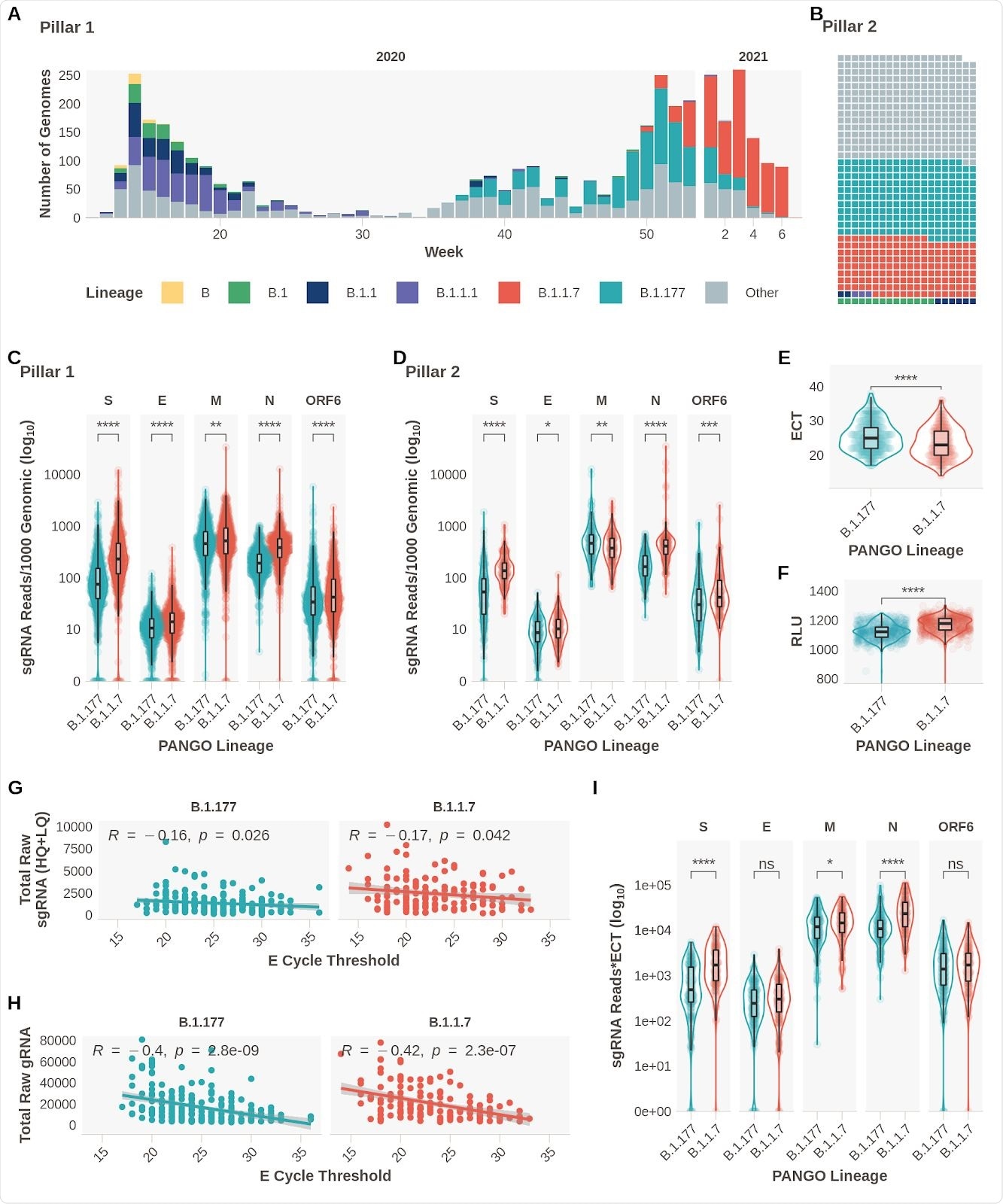
Changes observed in subgenomic RNA expression
The results confirmed the team’s hypothesis as they observed increased expression of subgenomic RNA of S, N, E, ORF6, and M proteins in patients with B.1.1.7 infection.
Evidence confirming previous findings of decreased E gene Ct (ECT) and increased relative light units (RLT) in patients infected with the B.1.1.7 variant were also noted.
There was also a weak but significant negative correlation between sgRNA reads and ECT. Also, there was a greater negative association between genomic reads and ECT.
ECT may have acted as a surrogate of viral load, explaining the rise in sgRNA expression in the B.1.1.7 variant. However, when the researchers reanalyzed the data while controlling for sgRNA to ECT, they found the changes were independent of ECT. They observed differences for S, N, and M sgRNA between the B.1.1.7 and B.1.177 variant.
Ruling out confounds
A potential explanation regarding sgRNA changes during infection could be because of symptom differences.
“It is possible that the days since symptom onset at sampling may vary between lineages in our dataset, either due to changes in sampling practice over time or presentation of individuals to healthcare services, which in turn could confound our sgRNA findings.”
To rule this out, the researchers collected symptom information from 2,327 positive SARS-CoV-2 samples. They found small differences when symptoms appeared for a B.1.177 infection and a B.1.1.7 infection. However, symptom duration was unlikely to affect the overall results. When controlling for these differences, the elevated subgenomic RNA levels for S, N, M, and E persisted.
The team also used a pseudo-infection to look at the timing of genetic changes days after symptom onset for both variants. They found infections with the B.1.1.7 variant showed increased sgRNA expression with an early peak on the first day of symptom onset. The researchers suggest ACE2 and TMPRSS2 expression may have expedited sgRNA kinetics, enhancing sgRNA expression early on in the infection.
When the team analyzed noncanonical sgRNA expression, they found decreases in the templates for all samples. Samples containing the B.1.1.7 variant showed greater reads, which the researchers suggest supports the production of noncanonical sgRNA from the genomic position 28282.
The B.1.1.7 variant has a triple nucleotide sequence containing 28282 in their genomic sequence, causing the D3L amino acid substitution in the nucleocapsid protein.
The researchers suggest the N D3L mutation may have been caused by recombination between noncanonical sgRNA and the genome. The result could have produced significant enrichment of sgRNA containing ORF9b.
This coincides with the 16-fold expression of ORF9b sgRNA observed in B.1.1.7 samples. In contrast, sgRNA-containing ORF9b was found in only 13.8% of non-B.1.1.7 samples. Additionally, non-B.1.1.7 samples did not contain ORF9b sgRNA with the N D3L mutation.
Understanding variants of concern
Based on the findings, the team proposes that sgRNA-containing ORF9b could be mediating interferon responses exhibited during SARS-CoV-2 infections. This suggests the sgRNA containing ORF9 could be causing greater infectiousness in the B.1.17 variant, but this should be explored further in future SARS-CoV-2 experiments.
They recommend using data on sgRNA for all genomic surveillance efforts to analyze the effects of SARS-CoV-2 variation and it affects the viral genome.
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Parker MD, et al. Altered Sub-Genomic RNA Expression in SARS-CoV-2 B.1.1.7 Infections. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.03.02.433156, https://www.biorxiv.org/content/10.1101/2021.03.02.433156v2
Posted in: Medical Research News | Disease/Infection News
Tags: ACE2, Amino Acid, Coronavirus, Coronavirus Disease COVID-19, CT, Gene, Genetic, Genome, Genomic, Genomic Sequencing, Healthcare, Mutation, Nucleotide, Protein, Public Health, Research, Respiratory, RNA, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Throat

Written by
Jocelyn Solis-Moreira
Jocelyn Solis-Moreira graduated with a Bachelor's in Integrative Neuroscience, where she then pursued graduate research looking at the long-term effects of adolescent binge drinking on the brain's neurochemistry in adulthood.
Source: Read Full Article