eating too much ibuprofen

A study conducted by researchers in Germany has found that the B.1.617 variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that has emerged in India entered certain types of lung and intestine cells with slightly increased efficiency compared with the original wild-type strain.
The B.1.617 variant is the lineage thought to be responsible for the sharp rise in coronavirus disease 2019 (COVID-19) cases and deaths in India over recent weeks.
The team – from the German Primate Center in Göttingen, the University of Göttingen Medical Center, the Friedrich-Alexander University of Erlangen-Nürnberg and Hannover Medical School – also reports that the entry of B.1.617 into lung and intestinal cells was blocked following treatment with soluble angiotensin-converting enzyme 2 (ACE2) or the serine protease inhibitor Camostat.
However, this host cell entry was not blocked by the monoclonal antibody Bamlanivimab, which has received emergency use authorization (EUA) as a COVID-19 treatment.
Finally, B.1.617 also partially evaded neutralization by the antibodies induced through natural infection or immunization with the Pfizer-BioNTech BNT162b2 vaccine.
Markus Hoffmann and colleagues say that antibody evasion by B.1.617 may contribute to the rapid spread of this variant.
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.
.jpg)
The emergence of variants threatens efforts to contain the pandemic
Since the COVID-19 outbreak first began in late December 2019, imitrex injection onset of action variants of the causative agent SARS-CoV-2 have emerged that threaten efforts to contain the pandemic.
For example, the B.1.1.7 lineage that emerged in the UK and is now spreading in many countries shows increased transmissibility compared with previously circulating strains.
Researchers suspect that this increased transmissibility may be linked to the N501Y mutation in the receptor-binding domain (RBD) of the viral spike protein – the main structure the virus uses to infect host cells.
This spike protein is comprised of two subunits. The RBD in subunit 1 binds to the host cell receptor ACE2, which is followed by activation of the spike by transmembrane protease serine 2 (TMPRSS2) or other cellular proteases. Subunit 2 (S2) then facilitates the fusion of the virus and cell membrane to enable the delivery of the viral genome into the cell.
These processes are essential for SARS-CoV-2 infection, and the spike protein is the primary target of monoclonal antibody therapies and the neutralizing antibodies generated following vaccination or natural infection.
What about the other variants?
The B.1.351 and P.1 variants that became dominant in South Africa and Brazil, respectively, harbor the spike RBD mutation E484K, which has been shown to reduce neutralization by antibodies.
So far, antibody evasion has been shown to be most prominent for variant B.1.351, however, it is unclear whether variants may arise that exhibit yet further or even complete resistance to neutralization.
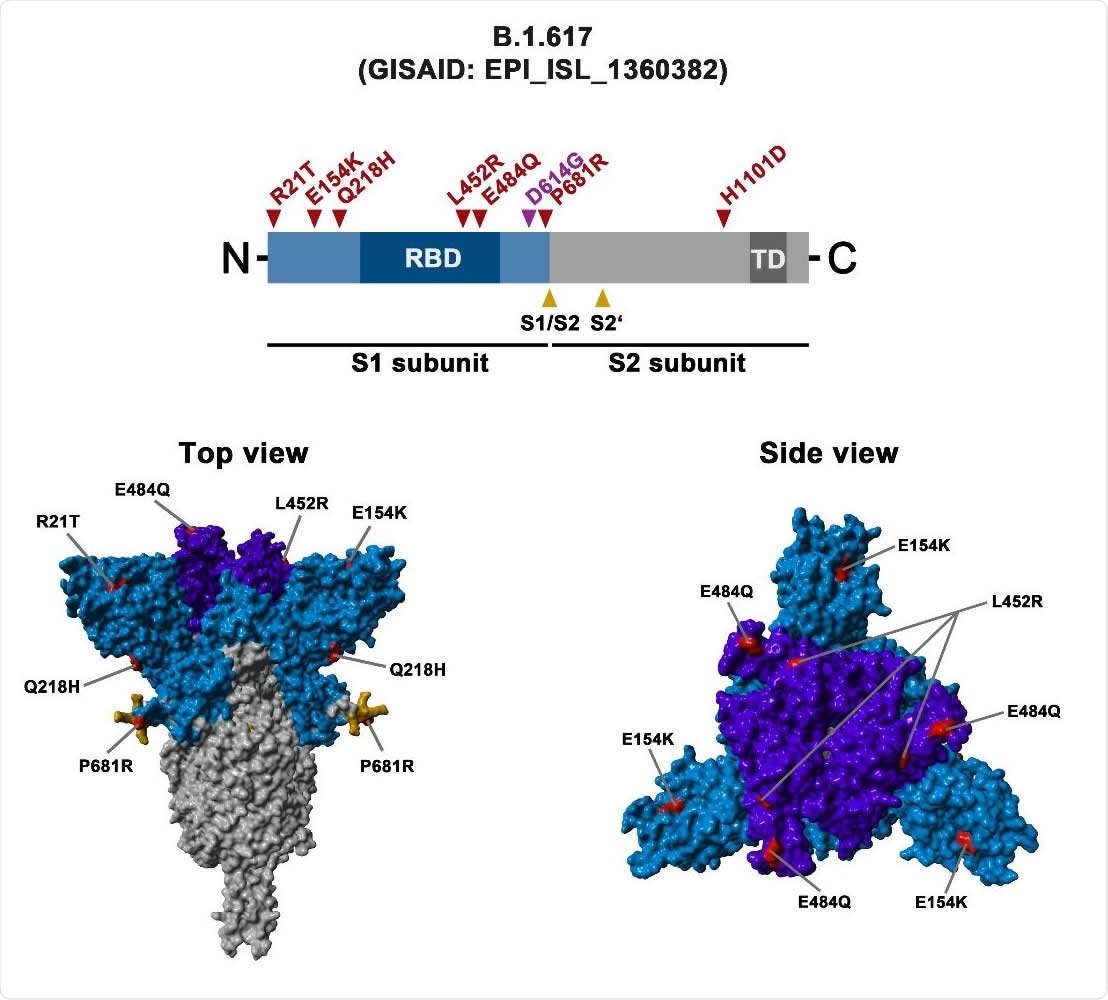
The sharp rise in COVID-19 cases and deaths in India over recent weeks is thought to be caused by the novel variant B.1.617, which harbors eight mutations in the spike protein. These include the RBD mutations L452R and E484Q, which are known to modulate antibody-mediated neutralization.
However, the researchers say it is not currently known whether B.1.617 resists this antibody-mediated neutralization.
The researchers say it is also unknown whether these spike mutations might alter important properties such as the efficiency of host cell entry or susceptibility to certain drugs.
What did the researchers do?
To test whether B.1.617 is more adept at entering cells, the researchers infected eight cell lines with pseudotyped virus particles expressing spike protein from the original wild-type virus, the B.1.617 variant, or the B.1.351 variant.
Of the eight cell lines tested, the spikes of B.1.617 and B.1351 mediated entry into Calu-3 lung cells and Caco-2 colon cells with slightly increased efficiency, compared with the original strain.
“The spike protein of SARS-CoV-2 B.1.617 allows for moderately enhanced entry into certain cells of the respiratory and digestive tracts,” says Hoffman and colleagues.
Next, the researchers showed that this B.1.617 spike-driven entry could be inhibited by soluble ACE2, which targets the RBD and blocks subsequent engagement of membrane-bound ACE2. Cellular entry was also blocked by the clinically proven serine protease inhibitor Camostat, which targets TMPRSS2.
“These results indicate that soluble ACE2 and Camostat will be active against the B.1617 variant,” writes the team.
Neutralization by monoclonal antibodies, convalescent and vaccinated sera was affected
By contrast, cellular entry of B.1.617 was partially resistant to neutralization by Casirivimab and fully resistant to neutralization by Bamlanivimab – two monoclonal antibodies that have received emergency use authorization as COVID-19 treatments.
“These results suggest that Casirivimab and particularly Bamlanivimab monotherapy may not be suitable for the treatment of patients infected with variant B1.617,” say the researchers.
Furthermore, B.1.617 cellular entry was partially resistant to neutralization by sera from convalescent individuals and sera from individuals immunized with two doses of the Pfizer BNT162b2 vaccine.
Antibody evasion by B.1.617 may contribute to its rapid spread
“The present study reveals that the B.1.617 spike protein can facilitate entry into Calu-3 lung and Caco-2 colon cells with slightly increased efficiency and shows that entry can be blocked by soluble ACE2 and Camostat,” says Hoffman and colleagues.
“In contrast, Bamlanivimab, a recombinant antibody with EUA did not inhibit entry driven by the B.1.617 spike and evidence for moderate evasion of antibodies induced by infection, and BNT162b2 vaccination was obtained,” they add.
Collectively, the study reveals that antibody evasion by B.1.617 may contribute to the rapid spread of this variant, concludes the team.
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Hoffman M, et al. SARS-CoV-2 variant B.1.617 is resistant to Bamlanivimab and evades antibodies induced by infection and vaccination. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.05.04.442663, https://www.biorxiv.org/content/10.1101/2021.05.04.442663v1
Posted in: Medical Research News | Disease/Infection News
Tags: ACE2, Amino Acid, Angiotensin, Angiotensin-Converting Enzyme 2, Antibodies, Antibody, Cell, Cell Membrane, Coronavirus, Coronavirus Disease COVID-19, Drugs, Enzyme, Genome, Immunization, Medical School, Monoclonal Antibody, Mutation, Pandemic, Protein, Receptor, Research, Respiratory, SARS, SARS-CoV-2, Serine, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Spike Protein, Syndrome, Vaccine, Virus

Written by
Sally Robertson
Sally first developed an interest in medical communications when she took on the role of Journal Development Editor for BioMed Central (BMC), after having graduated with a degree in biomedical science from Greenwich University.
Source: Read Full Article