Researchers develop model for SARS-CoV-2 fusion intermediate that helps membrane fusion

A model of the fusion intermediate shows that it is highly flexible and can undergo significant changes in its conformation to look for host membranes to capture.
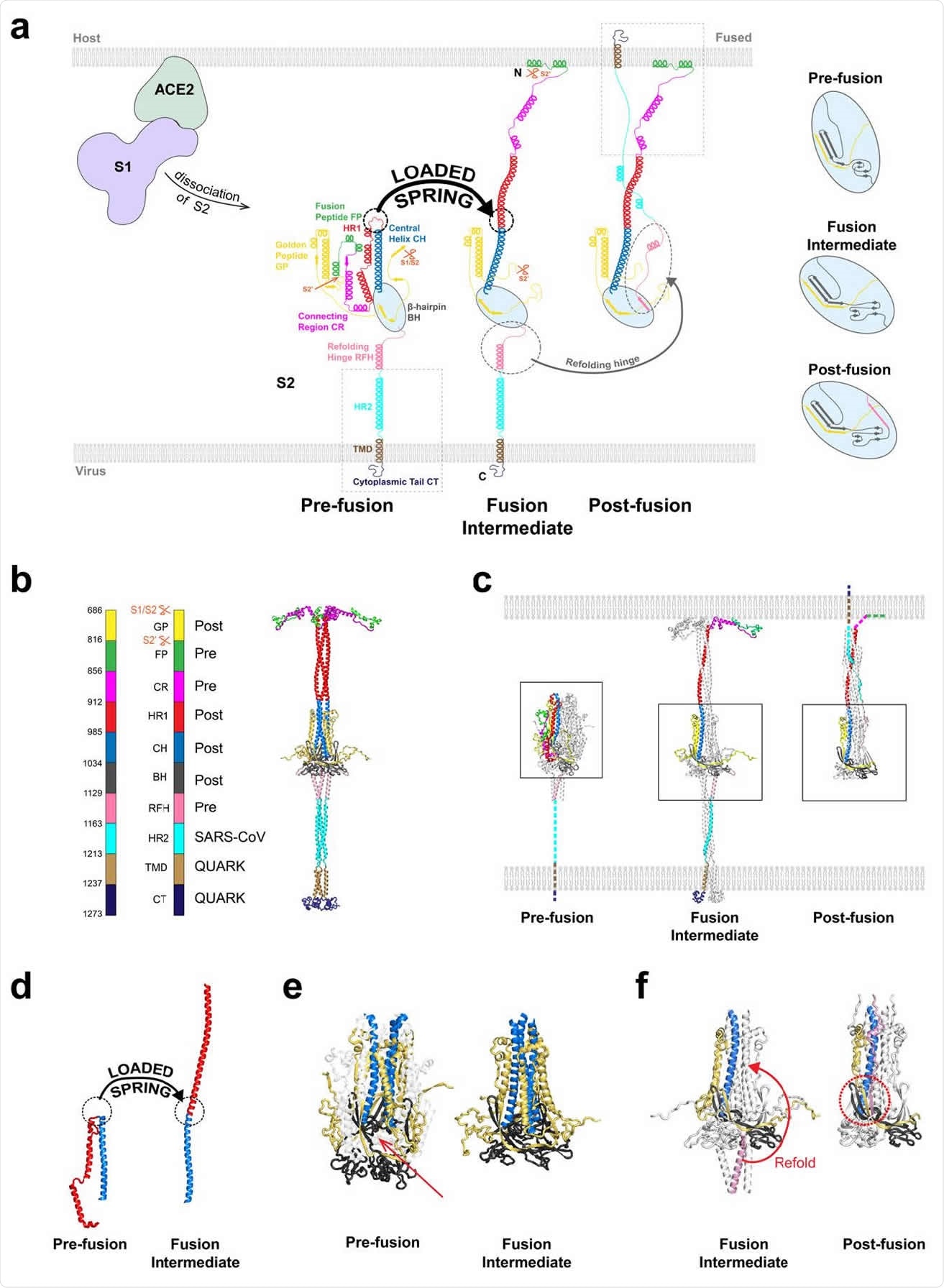
The spike protein is the main part of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) responsible for the infection. It consists of two subunits S1 and S2. Upon entry of the virus into the human body, the S1 subunit helps the virus bind to the angiotensin-converting enzyme 2 (ACE2) receptors on host cells.
After binding, the S1 and S2 subunits are thought to dissociate, releasing S2, which helps membrane fusion of the virus to the host. However, before this can happen, S2 needs to change its structure to the fusion intermediate (FI) form to allow membrane fusion.
The FI has three N-terminal fusion peptides, which are inserted into the host cell membrane. Then the membranes are refolded, helping fusion, and the virus genome enters via fusion pore.
However, not much is known about the FI, in particular, its structure. One challenge is the short lifetimes of the FI. The lack of experimental data makes it difficult to do any computational studies of the intermediate. A better understanding of how the fusion process works in SARS-CoV-2 may help develop better therapeutics to combat emerging variants.
A model for the fusion intermediate
With this aim, researchers from Columbia University in New York constructed a model for the SARS-CoV-2 FI using available data and computationally explored its behavior. The research is published on the bioRxiv* preprint server prior to peer review.
Based on how the influenza hemagglutinin, another viral fusion protein, behaves, the team thought the transition to the FI starts with changing all the heptad repeat 1 (HR1) domains into alpha-helices, so that the unstructured loops in HR1 become helical, like springs, becoming continuous with the central helix.
Using this and the structures of other components, the team came up with a model for FI. They tested their model using molecular dynamics simulations and found it to be stable. They found the regions below the β-hairpin domain to be highly flexible, allowing the FI to tilt.
The prefusion state likely changes to the FI state by a loaded spring release mechanism, where the loop-to-helix transitions become straight, extending the HR1 domain. Simulations of the fusion peptide determined an N-terminal helix with 10 residues that first contacts the membrane.
The author posits that the spring-loaded release occurs first. Then any refolding of the refolding hinge domain occurs after a delay, which allows the FI to change conformation to destabilize a refolding hinge domain into an unstructured loop.
Kinetics of the fusion intermediate
When the authors ran simulations to capture the dynamics of the FI, they found the FI showed significant configuration changes, sweeping a volume of about 25,000 nm3. They identified three hinge regions at the base of the FI, which allows the FI to be highly flexible so that it can cover a large volume of space looking for the membrane for fusion.
The hinges could have several important roles. They increase the accessible area available to the fusion peptides on the FI terminal, capturing the host cell membrane. The flexibility of the FI may also allow many FIs to capture the membrane at different locations and present the fusion peptides directly to the target membranes.
The researchers also found that secondary structures at the base of the FI were also highly dynamic. The three refolding hinge domain helices were spread out like an inverted tripod, buffering the longitudinal changes in the FI backbone. The spread-out helices also allowed upstream refolding hinge domains more flexibility.
Studies that have used isolated fusion peptides show a much rapid binding rate than seen in the FI model. Compared to what previous studies predict, about a 15 μs delay time for the virus to capture the membrane, the researchers did not see any membrane capture up to about 300 μs. This suggests binding of the fusion peptides is much slower when they are attached to the FI.
In addition, the team found that the golden peptide, a disconnected fragment between the S1/S2 site and the S2 site, may chaperone refolding. Monoclonal antibodies that target a region in the refolding hinge helix, which engages the golden peptide, have been seen before. This suggests the golden peptide or the refolding hinge may be potential targets for developing therapeutics against COVID-19. Unveiling the mechanisms governing the CoV-2 S2 fusion intermediate will play a vital role in the search for robust and pan-coronavirus antiviral drugs.
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Su, R. et al. (2021) Large fluctuations of the fusion intermediate help SARS-CoV-2 capture host cell membranes. bioRxiv, https://doi.org/10.1101/2021.04.09.439051, https://www.biorxiv.org/content/10.1101/2021.04.09.439051v2
Posted in: Medical Research News | Disease/Infection News
Tags: ACE2, Angiotensin, Angiotensin-Converting Enzyme 2, Antibodies, Cell, Cell Membrane, Chaperones, CLARITY, Coronavirus, Coronavirus Disease COVID-19, CT, Drugs, Enzyme, Genome, Helix, Influenza, Peptides, Protein, Research, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Spike Protein, Syndrome, Therapeutics, Virus

Written by
Lakshmi Supriya
Lakshmi Supriya got her BSc in Industrial Chemistry from IIT Kharagpur (India) and a Ph.D. in Polymer Science and Engineering from Virginia Tech (USA).
Source: Read Full Article