methylphenidate dea schedule
<img class="aligncenter" src="https://scx1.b-cdn.net/csz/news/800a/2023/seeking-vulnerabilitie.jpg"
alt="Seeking vulnerabilities within SARS-CoV-2"
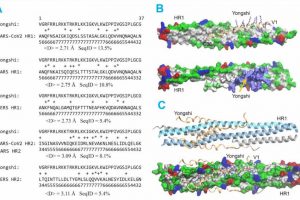
title="Deep-learning sequence alignment algorithm and computational modeling predict stable binding interactions between Yongshi and HR1. (A) Sequence alignments between Yongshi and HR1/HR2 sequences of SARS-CoV-1, SARS-CoV-2, and MERS-CoV by SAdLSA . denotes the mean of predicted distance between aligned residues, and SeqID is Sequence Identity. Identical and statistically favorable amino acid substitutions are denoted by "*" and "+", respectively. The digits below each alignment indicate the probability that the predicted structure for each aligned amino acid pair will be less than 3 Å, tetracycline hcl skin ointment e.g., the number 6 predicts a probability score between 60 to 70%. Note that SARS-CoV-1 and SARS-CoV-2 have identical sequences in their HR2 regions aligned to Yongshi. (B) A structural model of two SARS-CoV-2 HR1s in complex with a single Yongshi peptide. The two HR1s are shown in the surface representation, and the Yongshi peptide is shown in ribbon backbone and licorice sidechain representations in the upper snapshot, and the same surface representation in the lower snapshot. The lower snapshot is slightly rotated from the upper snapshot to show a large positively charged surface patch of Yongshi as pointed by a yellow arrow. The color codes for amino acids are hydrophobic (white), polar (green), positive (blue), negative (red). (C) A structural model of three SARS-CoV-2 HR1s in complex with three Yongshi peptides after 100 ns of MD simulation. The upper snapshot is shown in cartoon representation for three HR1s (blue, cyan, gray), and ribbon representation for Yongshi (yellow). The lower snapshot is in the same orientation as the upper one but shown in the same representation as snapshots in (B). Credit: Scientific Reports (2023). DOI: 10.1038/s41598-023-41850-7″ width=”800″ height=”509″>
Emory Vaccine Center researchers say they have identified the Achilles heel of SARS-CoV-2, the virus that causes COVID-19.
Part of the viral fusion machinery seems to stay the same among several viral variants, and this vulnerable machinery is what the antiviral peptide Yongshi targets. A peptide is a small protein or fragment of a protein.
Derived from a natural antiviral peptide produced by wild boar, Yongshi (meaning “warrior” in Mandarin) can prevent SARS-CoV-2 from infecting host cells. The scientists are now studying how to adapt the peptide for clinical use. Their research is now published in Scientific Reports.
“Yongshi throws a wrench into the machine that allows SARS-CoV-2 to enter the host cell,” says senior author Joshy Jacob, Ph.D., professor of microbiology and immunology at Emory University School of Medicine. Jacob’s Vaccine Center lab is based at Emory National Primate Research Center.
An advantage for Yongshi is that it appears to be active against all known SARS-CoV-2 variants, including the recent XBB.1.16. It is also active against the related viruses SARS-CoV-1 and MERS. For comparison, monoclonal antibodies developed against SARS-CoV-2 variants have often lost activity as the virus continues to evolve and new variants emerge.
“This is important because drug makers and the public in general has to catch up whenever a new viral variant takes the stage,” Jacob says. “A peptide-based antiviral approach might allow us to stay ahead.”
The origin of Yongshi was in cathelicidin peptides, which are produced by mammals, birds, amphibians and fish and have long been studied for their antimicrobial properties.
Working in Jacob’s lab, former graduate student Troy von Beck tested cathelicidin peptides from several types of animals, including bats, pangolins, humans, whales, snakes, cats, koalas and wallabies. He settled on one from wild boar, which was the most active, and then modified it to reduce toxicity against human cells. Pigs are not susceptible to SARS-CoV2, and it is unclear whether their cathelicidin peptides may be partly responsible.
To understand the mechanism of Yongshi’s antiviral activity, the Emory researchers collaborated with computational protein-modeling experts Jeffrey Skolnick, Ph.D., and Mu Gao, Ph.D., at Georgia Tech.
In a computer simulation, they identified interactions between Yongshi and part of the SARS-CoV-2 spike protein known as the “helical bundle.” Further experiments with the Emory-based labs of Mehul S. Suthar, Ph.D., and Renhao Li, Ph.D., supported this model. Previously discovered helical bundle peptide inhibitors are expected to be cross-reactive with SARS-CoV-2 antibodies and may be ineffective in vaccinated or convalescent individuals.
Jacob’s lab has a track record of finding antiviral peptides from the natural world. In collaboration with Indian herpetologists, his team isolated an anti-flu peptide from frog skin secretions. They later found another peptide from frogs against Zika and dengue viruses.
Developing antimicrobial peptides into effective drugs has been a challenge, partly because enzymes in the body can break them down.
Jacob says that further modifications to the peptide are necessary to generate a therapeutically viable product. This may include chemical modification, which could both reduce the necessary dose of peptide and improve its stability following administration. Also, Jacob suggests that mRNA technology used in vaccines against SARS-CoV-2 vaccines could be a way to deliver Yongshi to patients long-term, either as a single-dose therapeutic following infection or as a prophylactic prior to infection.
More information:
Troy von Beck et al, A wild boar cathelicidin peptide derivative inhibits severe acute respiratory syndrome coronavirus-2 and its drifted variants, Scientific Reports (2023). DOI: 10.1038/s41598-023-41850-7
Journal information:
Scientific Reports
Source: Read Full Article