Pangolin coronavirus sheds light on UK SARS-CoV-2 variant's evolutionary trajectory

Researchers at the Division of Infection and Immunity, University College London, UK, have provided key insights into the characteristics of the B.1.1.7 (or UK) variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
The team report that the B.1.1.7 variant, first identified in the United Kingdom in September 2020, appears to have several functional mutations that aid in cell entry. Additionally, the virus appears to have similar changes in the spike protein to pangolin coronaviruses – these changes also allow pangolin coronavirus to readily enter human cells, suggesting future pathways for SARS-CoV-2 to evolve for more efficient cell entry.

A pre-print version of the research paper is available to read in full on the bioRxiv*server.
Variants of concern
SARS-CoV-2 is the causative agent of the coronavirus disease 2019 (COVID-19) pandemic that is now circulating globally. The origins of this virus are presently unknown, but are presumed to be zoonotic in nature, likely originating from either bat or pangolin-related coronaviruses.
Presently, three variants of concern have been identified and are under surveillance: B.1.1.7 originating in the UK, B.1.351 from South Africa, and P.1 from Brazil. These variant lineages are all associated with a number of convergent mutations that promote higher transmissibility, antibody evasion, and/or more efficient cell entry.
The B.1.1.7 variant contains mutations that allow for all three of the prior mentioned advantages, and as such, is of great concern as it continues to become more prevalent in numerous countries.
The researchers in this study sought to investigate the role of these mutations, specifically how they differ from the ancestral Wuhan-Hu-1 virus and related pangolin viruses, which shared 97% similarity on the spike protein responsible for mediating cell entry.
The study – part 1
SARS-CoV-2 binds to the angiotensin-converting enzyme 2 (ACE2) receptors on the cell membrane of principally respiratory and cardiac cells.
The researchers first compared the efficiency of cell entry of pseudoviruses containing the B.1.1.7 spike with the Wuhan-Hu-1 (ancestral) reference strain spike. They observed that in three model cell lines all commonly exploited by SARS-CoV-2 (HeLa ACE2, Calu-3, and HEK 293T), B.1.1.7 showed greater entry to all of these than Wu-Hu-1, particularly so in HEK 293T (~10 fold enhancement versus a ~3 fold enhancement in the remaining two).
HEK 293T cells have a very low expression of ACE2, roughly 500 times lower than in HeLA ACE2 and Calu-3 cells. The researchers conclude from this that while the B.1.1.7 spike retains the same mode of entry as the ancestral spike – through ACE2 receptor binding – it also has mutations that allow for much greater infection in suboptimal conditions.
The study – part 2
Secondly, the researchers compared the efficiency of cell entry between Wu-Hu-1, B.1.1.7, and a modified Wu-Hu-1 spike bearing the D614G mutation. This mutation is convergently found in all three variants of concern, and is thought to promote cell entry (although this seems to also partially decrease antibody resistance).
They observed that Wu-Hu-1 D614G viruses saw greater infection in HeLa ACE2 cells than Wu-Hu-1 and B.1.1.7, additionally that Wu-Hu-1 D614G exhibited equal activity as B.1.1.7 in HEK 293T cells. From this, they could conclude that additional mutations pertained by B.1.1.7 limit cell entry for the virus in some cell types but maintains it in others.
The B.1.1.7 virus also has several deletion mutations in the N-terminal domain (NTD). The researchers replicated these deletion mutations in the Wu-Hu-1 spike, and restored these in the B.1.1.7 spike. Wu-Hu-1 spikes with NTD deletions were indistinguishable from their origin spike. However, the B.1.1.7 entry phenotype was eradicated in the restored spike. This implies that the NTD plays a currently unknown role in protein activity, which the authors say warrants future investigation.
The study – part 3
With SARS-CoV-2’s presumed zoonotic origin, the research team compared the cell entry abilities of bat (RaTG13) and pangolin coronaviruses (GD – Guangdong isolate) into human cells. RaTG13 spike is ~97% similar to that of SARS-CoV-2, and the receptor-binding domain (RBD) of Pangolin CoV GD is ~97% similar to that of SARS-CoV-2.
The team observed minimal infection by RaTG13, as seen in previous studies, due to poor binding ability. However, Pangolin CoV showed high affinity for human ACE2, having a 50-100 fold greater infection of HeLa ACE2 and HEK 293T than SARS-CoV-2. Pangolin CoV also exhibited similar levels of antibody evasion to B.1.1.7, suggesting similar evasion mechanisms.
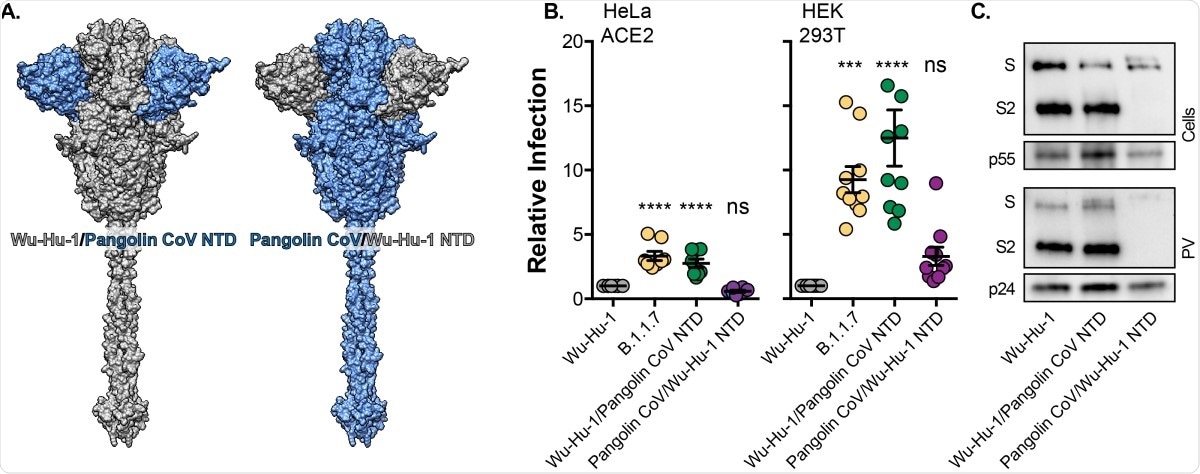
The study – part 4
Finally, genetic swaps of NTD deletions in the spikes of Wu-Hu-1 and Pangolin CoV were observed. Interestingly, the Wu-Hu-1/Pangolin CoV NTD yielded the same infection results as B.1.1.7 on the three cell models, while the Pangolin CoV/Wu-Hu-1 NTD was indistinguishable from Wu-Hu-1. This further highlighted the likelihood that presently NTD mutations play a large role in facilitating cell entry and invasion.
Concluding remarks
The authors highlight the role that the NTD region of SARS-CoV-2 currently plays. Understanding mutations in this region may be crucial in anticipating future cell entry- or antibody evading adaptations.
*Important notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Dicken S., et al. Characterisation of B.1.1.7 and Pangolin coronavirus spike provides insights on the evolutionary trajectory of SARS-CoV-2. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.03.22.436468, https://www.biorxiv.org/content/10.1101/2021.03.22.436468v1
Posted in: Medical Science News | Medical Research News | Disease/Infection News | Healthcare News
Tags: ACE2, Angiotensin, Angiotensin-Converting Enzyme 2, Antibody, Cell, Cell Membrane, Coronavirus, Coronavirus Disease COVID-19, Enzyme, Genetic, Mutation, Pandemic, Phenotype, Phylogeny, Protein, Receptor, Research, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Spike Protein, Syndrome, Virus, Zoonosis

Written by
Michael Burgess
Michael graduated with a first-class degree in Zoology from the University of Hull, and later received a Masters degree in Palaeobiology from the University of Bristol.
Source: Read Full Article