Automated Zebrafish screening for Myeloid Leukemia drug discovery
 Thought LeadersDr Elspeth Payne & Dr Alexandra LubinUCL Cancer Institute
Thought LeadersDr Elspeth Payne & Dr Alexandra LubinUCL Cancer InstituteIn this interview, NewsMedical speaks to Elspeth Payne and Alexandra Lubin at UCL Cancer Institute, who explain how they use zebrafish modeling to study the development of myeloid leukemias.
Could you please introduce yourself, what it is you do and briefly describe your research?
Elspeth Payne: My name is Beth Payne and I am a clinician scientist. I work on myeloid disorders in the laboratory and look after patients in the hospital; my laboratory works on blood cancers, particularly myeloid malignancies. We use a combination of zebrafish modeling and drug screens alongside primary patient material to try and study better ways to understand the development of myeloid leukemias and different ways that we could treat them in the future.
Alexandra Lubin: My name is Alex Lubin. I am a post-doctoral research scientist in Beth Payne's lab. I work on zebrafish models of myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML). Drug screening is my main project and I am looking at doing some drug screening in hematopoietic stem cells in the tails of zebrafish to look for synthetic lethality.
How did you perform analysis of zebrafish before using the Athena software?
Alexandra Lubin: When I am looking at stem cells in my drug screen, I try to find a change in number. To do this manually, all the individual fluorescent cells must be counted in the tail. That means you have to take each fish under a microscope, go through and count them, which is very time-consuming. For this reason, we started looking for an automated approach.
What are some of the challenges you face in getting the data you need?
Beth Payne: We have been developing some drug screening methodologies, and while the zebrafish is a great model for this, getting the right throughput has been challenging. Although there are many different platforms that one can use, they tend to have kind of bespoke solutions for how you analyze the data. That means it is costly and takes a long time to develop a process that works for your purpose.
At what point then did you move away from manual imaging and image analysis to search for automated solutions?
Beth Payne: We still use a lot of manual imaging as there are some situations that require its use. In terms of automated screening, we have been looking for a solution for probably the last five to ten years.
What are the main challenges when sampling with a manual workflow?
Alexandra Lubin: The main challenge with the manual method is that it is time-consuming. It is not particularly difficult to do, but it involves spending hours and hours at microscopes. The other main problem with that is you cannot achieve large sample numbers. When you want to run something like a drug screen, the power comes from being able to analyze many samples.

Image credit: IDEA Bio-Medical
What has this technology brought to your lab?
Beth Payne: The thing that has been helpful for this system is we use it for all indications rather than just drug screening. For example, if we have a transgenic or even an in-situ that we want to image and look at that in detail in high content, we will use the Athena rather than doing it manually.
The other great thing about the system is that it is flexible and you can optimize it for a different assay relatively quickly as the system is user-friendly. The company offers a lot of support to try and help us when we run into some difficulties.
I would say that overall it allows us to power our experiments much better because instead of using just 20 embryos, we can use 200 for the same processing time. Overall, it offers more flexibility.
Azo: When you started working on the Athena software, how long did it take you to be independent and productive with it?
Alexandra Lubin: When I first encountered the Athena software, it was very much in development. That gave room for many discussions with IDEA Bio-Medical about what we wanted the software to do, what would be useful, and highlighting areas for improvement.
Once it was up and running, the transition time from first using the software to becoming fully independent with it was relatively short.
How does that compare with other instrumentation you use, for example compared with manual microscopes?
Alexandra Lubin: This has been a unique experience in the sense that we have had more contact with IDEA Bio-Medical than we would usually have. I have never used another system like this, so I do not really have a direct comparison, but it is an excellent and extremely practical piece of software.
How long does it take you to analyze all your fish samples and how does that compare previously?
Alexandra Lubin: When conducting these experiments manually, we can only screen about a hundred fish at a time. However, that takes such a significant amount of time, but once we began using the Athena, we can increase that number.
How many fish I screen depends on how well-behaved the fish are, as anybody who works with zebrafish will know. It is possible to screen hundreds (of fish) in a short space of time as it only takes around 30 minutes between loading the plate and getting the results for about a hundred fish, whereas previously, that would have taken a whole day.
What do you do when you need some support using the software? Who supports you and how?
Alexandra Lubin: When I need some support with the software, I reach out to the team at IDEA Bio-Medical. They have been really helpful and offer a collaborative partnership. I provide them with images; they provide us with support – it works seamlessly.
What was the point when you thought the Athena software was useful to your research?
Alexandra Lubin: When I started this drug screen, we knew we needed to bring in automation as it was not going to be possible to screen thousands of compounds manually. When we started looking at different methods, it became clear how easy to use the system was and how flexible it was. Moreover, we were drawn to the fact that Athena enabled us to branch out and change it if we needed to.
Where do you see your work going next and the impact?
Beth Payne: I believe that it has certainly opened the door to performing more and more screens. Perhaps we may now be able to conduct more ambitious types of screens where we might use, for example, more than one fluorophore or more than one genotype at the same time because the time taken to screen is so much shorter and more efficient that you can introduce more variables.

About Dr Elspeth Payne
Dr Elspeth Payne is a Senior Clinical Researcher/Clinical Consultant UCL Cancer Institute. Her laboratory at UCL’s Cancer Institute is dedicated to the study of inherited bone marrow failure disorders and leukemias, and uses zebrafish to model these diseases. Dr Payne is also a clinical hematologist at UCL Hospital, where she treats people with blood disorders including leukemia and bone marrow failure.
 About Dr Alexandra Lubin
About Dr Alexandra Lubin
Dr. Lubin is a post-doctoral at the UCL Cancer Institute where she uses zebrafish to study myelodysplastic syndrome (MDS) and acute myeloid leukaemia (AML), aiming to develop novel therapeutic treatments. Previously, she obtained her PhD in Chemical Biology from Imperial College London after studying Chemistry at the University of Cambridge.
About IDEA Bio-Medical Ltd.
 IDEA Bio-Medical is founded in 2007 through a partnership between YEDA (the Weizmann Institute’s commercialization arm) and IDEA Machine Development (an innovation hub).
IDEA Bio-Medical is founded in 2007 through a partnership between YEDA (the Weizmann Institute’s commercialization arm) and IDEA Machine Development (an innovation hub).
IDEA Bio-Medical is a company specializing in automated microscopy and image analysis for the life science researchers. IDEA’s products, The Hermes imaging system and Athena image analysis SW, have contributed to over 100 scientific publications in peer-reviewed magazines, globally supporting high impact science.
IDEA Bio-Medical focuses on empowering zebrafish researchers to provide them with a reliable, robust solution for automated and unbiased Zebrafish image analysis by applying company’s knowledge and specialties.
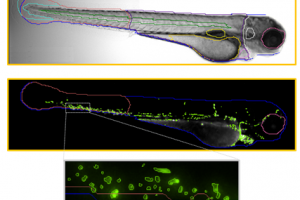
IDEA developed a novel deep learning-based image analysis software for in vivo zebrafish experiments. The software automatically detects zebrafish contour and their internal organs in brightfield with no required user input. The anatomy identified is coupled to fluorescence channels to permit anatomy-specific study of fluorescence changes. It is an affordable, user-friendly system designed specifically for reliable, automated zebrafish image-based analysis.
The software is suited for researchers who only image a handful of fish per week, as well as researchers imaging hundreds and thousands of fish in multi-well plates for large scale screens. As well, the software is available as a stand-alone product and accepts microscopy images in multiple image formats, including proprietary ones. So, all researchers using manual microscopes or automated systems from other vendors, can readily use IDEA’s Zfish software to extract quantitative, meaningful information from their Zebrafish images.
more info can be found here: https://idea-bio.com/products/zebrafish-image-analysis/
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.
Source: Read Full Article