New assay could revolutionize detection and treatment of acute myeloid leukemia
A novel assay that detects a unique molecular marker in patients with acute myeloid leukemia (AML) may revolutionize the way this disease is detected and treated according to a new report in The Journal of Molecular Diagnostics. This assay may improve the detection of AML driven by KMT2A gene fusions and may affect treatment decision-making, assessing response to therapy, and long-term surveillance.
AML is a rare, aggressive blood cancer diagnosed in around 120,000 individuals worldwide each year. Detecting residual disease during treatment is essential for determining prognosis and guiding treatment decisions. Currently, the methods for detecting measurable residual disease (MRD) during treatment for AML include bone marrow morphology, multiparameter flow cytometry (MPFC), and DNA sequencing.
Morphologic assessment only detects leukemic cells at a 5% limit of detection. MPFC has a more sensitive limit of detection at 0.01% to 0.001%, but is challenging to implement and interpret, and is not standardized between laboratories. DNA sequencing approaches can identify leukemic cells by their somatic mutation profile but are expensive and can be confounded by clonal hematopoiesis in non-leukemic blood cells.
Lead investigator Grant A. Challen, Ph.D., Division of Oncology, Department of Medicine, Washington University School of Medicine in St. Louis, explains, “Oncogenic fusions are often disease-defining and present a unique marker of leukemic cells that are not usually present in healthy cells. Other diseases, such as chronic myeloid leukemia (CML), can already be tracked by the canonical BCR-ABL fusion, and sensitively detecting these fusions has revolutionized how CML is treated.”
“For AML patients with oncogenic fusions driving their disease, the KMT2A fusion is a molecular marker that can be leveraged for sensitive MRD detection. We therefore wanted to develop a platform for sensitive KMT2A fusion-detection to improve how we detect and treat this disease.”
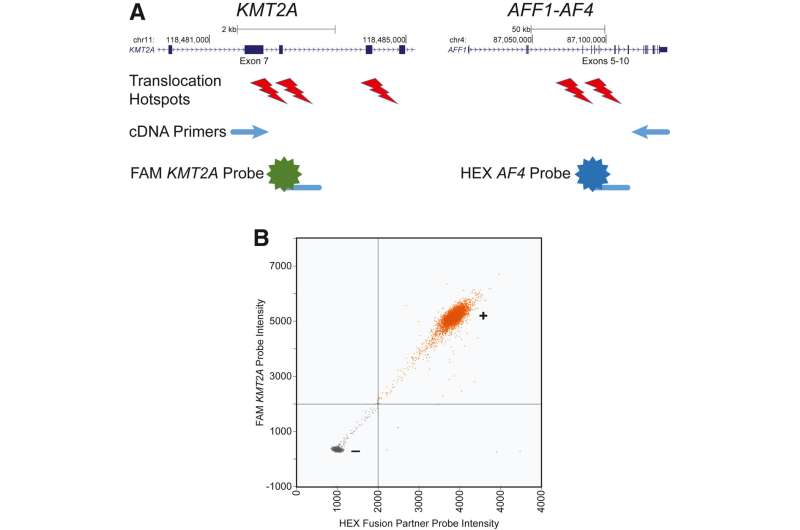
Investigators developed a novel droplet digital PCR assay enabling sensitive KMT2A fusion detection with the five most common fusion partners. There are at least 80 known KMT2A fusion partners, but about 80% of fusions involve just five partners—AF9, AF6, AF4, ELL, and ENL. They benchmarked the assay in human cell lines and patient samples to demonstrate sensitive and specific KMT2A fusion detection.
The assay detects these fusions by partitioning cDNA molecules into microfluidic droplets that are assayed with primers and probes that only produce a positive signal when fusion transcripts are present. Investigators were able to combine multiple primer/probe sets targeting different fusions into a pooled fusion detection reagent. They also showed the detection of KMT2A fusions in patient samples known to harbor KMT2A fusions.
Dr. Challen notes, “We show that the assay does not produce false-positive signals in samples from healthy individuals. The assay is easily expanded to include additional oncogenic fusions. This has potential impact for treatment decision-making and assessing response to therapy. Knowing whether a treatment is working or not is critically important for decisions regarding when to escalate therapy or pursue hematopoietic stem cell transplant.”
He concludes, “This is a robust new tool for sensitive KMT2A fusion detection that is directly applicable for disease detection in patients with leukemia driven by these fusions. It fills a void for oncogenic fusion detection and provides some technical improvements. The assay is also scalable—additional fusions can be easily added to the assay—to expand coverage for other oncogenic fusions. We are improving blood cancer detection one drop at a time!”
More information:
Andrew L. Young et al, Droplet Digital PCR for Oncogenic KMT2A Fusion Detection, The Journal of Molecular Diagnostics (2023). DOI: 10.1016/j.jmoldx.2023.09.006
Journal information:
Journal of Molecular Diagnostics
Source: Read Full Article