Notch Signaling Pathway

Notch signalling is involved in the cell-cell communication process via influencing the patterns of gene expression and differentiation. Notch pathway maintains stem cell populations and regulates binary cell fate choice. It also plays the role of mediator during short-range cell-cell communication. During development, this evolutionary conserved pathway functions in various cell types and stages.
In metazoans, Notch signalling is involved in cell proliferation, cell fate, cell differentiation, and cell death. Misregulation or loss of Notch signalling pathway is associated with several human disorders, including developmental syndromes, adult onset diseases, and cancer.
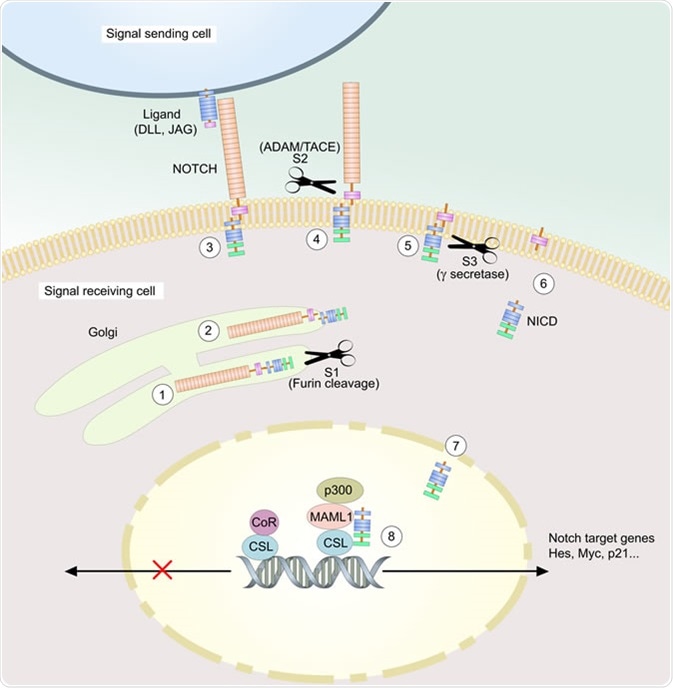
The Core Pathway
Notch is a single transmembrane protein located on the cell surface with transmembrane ligands. The signalling event is triggered by cell-cell contact which is essential for this event. Notch plays a dual role as a transmembrane receptor and a transcription factor.
Notch ligands stimulate the signalling pathway referred to as the canonical Notch signalling pathway. When a Notch transmembrane receptor establishes contact with a cell, it interacts extracellularly with the Notch transmembrane ligand which initiates proteolytic cleavage of the receptor and consequently the Notch intracellular domain (Notch ICD or NICD) of the receptor is released. Later Notch ICD interacts with CBF1/Suppressor of Hairless/LAG-1 (CSL) family of DNA-binding proteins after translocating to the nucleus. This DNA-binding protein is considered as the recombination signal binding protein specific to immunoglobulin kappa J region present in mammals. The lack of an amplification step is a feature of the core canonical Notch pathway.
Notch Ligand-Receptor Interactions
Mammals have four notch receptors and five canonical ligands. A huge number of receptor-ligand combinations are generated and are responsible for distinct responses. Notch receptors can also undergo post-translational modifications which modify the receptor-ligand interactions. Addition of O-glucose or O-frucose helps in the modification of notch receptors.
Notch Receptor Processing
Proteolytic processing of the Notch receptor is dependent on the activation of ligand. This results in the release of Notch ICD which translocates to the nucleus. Regulating the processing and relocalization events at multiple steps can further modulate Notch signalling.
Endocytosis and Trafficking of Processed Notch Receptors
Endocytosis is considered a crucial step in the transmission of the Notch signal. During endocytosis of Notch receptor, mono-ubiquitylation of the receptor is required at lysine 1749 and deubiquitylation is facilitated by elF3f.
The Notch Intracellular Domain – a Well-Decorated Signalling Hub
After traveling to the nucleus, the Notch receptor interacts with CSL which directs the transcription of downstream genes in the canonical Notch signalling pathway. Notch ICD goes through several post-translational modifications, including phosphorylation, ubiquitylation, hydroxylation, and acetylation.
Sources
- Cell and molecular biology of Notch (http://joe.endocrinology-journals.org/content/194/3/459.full.pdf)
- The Canonical Notch Signaling Pathway: Unfolding the Activation Mechanism (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2827930/)
- Notch Signaling (http://cshperspectives.cshlp.org/content/4/10/a011213)
- Notch signaling: simplicity in design, versatility in function (http://dev.biologists.org/content/138/17/3593#sec-3)
Further Reading
- All Cell Signaling Content
- The cAMP signaling pathway
- Phosphatidylinositol Biphosphate (PIP2) Signal Pathway
- GPCR Signaling Pathways
- GPCR Origin and Diversification
Last Updated: Feb 26, 2019

Written by
Amrita Roy
Amrita is a freelance science and medical writer from India. She has a B.Sc. in Microbiology from the University of Calcutta, and holds a post graduation degree in Microbiology. Amrita loves to travel to various places. She enjoys cooking and has her own food blog where she shares easy and tasty recipes.
Source: Read Full Article