Potent monoclonal antibody shows promise against emerging SARS-CoV-2 variants of concern

In a new study currently available on the bioRxiv* preprint server, a group of US-based researchers developed a panel of neutralizing monoclonal antibodies against SARS-CoV-2 that can block viral attachment to cells – and pinpointed a specific one that can potently neutralize all tested variants of concern.
.jpg)
As the coronavirus disease 2019 (COVID-19) pandemic is still actively unfolding, the ongoing vaccine effort should be supplemented with effective treatment strategies. Thus far, two treatments with monoclonal antibodies were approved for use in affected individuals under Emergency Use Authorization (EUA).
Previous studies have shown that most protective and potently neutralizing anti-SARS-CoV-2 monoclonal antibodies bind the receptor-binding domain (RBD) of the viral spike glycoprotein, albeit there are also some inhibitory monoclonal antibodies that target the N-terminal domain (NTD).
Consequently, selection pressure can result in SARS-CoV-2 selecting for mutations in the RBD and NTD that make possible the escape from antibody recognition and neutralization. This is the case with several emerging viral variants of concern that carry such mutations and, in turn, confer resistance to monoclonal or polyclonal antibodies elicited by natural infection or vaccines.
Hence, additional monoclonal antibodies or vaccines that preserve their efficacy against SARS-CoV-2 variants may be needed in our armamentarium in order to tackle all these new and evolving strains that we are faced with.
In this new paper, a research group led by Dr. Laura A. VanBlargan from the Washington University School of Medicine described a panel of very potent, neutralizing murine monoclonal antibodies against the RBD of SARS-CoV-2 that can bind several epitopes proximal to the receptor-binding motif of the RBD.
Binding and neutralization experiments
Here, the researchers have characterized a panel of anti-SARS-CoV-2 monoclonal antibodies, explained their mechanism of action on a cellular level, and also appraised in vitro neutralizing and in vivo protection capacity against historical and circulating SARS-CoV-2 variants.
The monoclonal antibodies were also tested for cross-reactive binding to the original SARS-CoV-1 spike protein. Furthermore, this research group has also ascertained the structure of the viral spike protein bound to SARS2-38 – a potently and broadly neutralizing monoclonal antibody that recognizes emerging variants of concern.
Neutralizing activity testing was pursued with the use of focus-reduction neutralization test (FRNT) and Vero E6 cells, with the letter representing kidney epithelial cells derived from an African green monkey. Epitope mapping of anti-SARS-CoV-2 monoclonal antibodies was also done with the use of neutralization escape analysis.
Monoclonal antibodies against variants of concern
While certain types of potently neutralizing monoclonal antibodies protected K18-hACE2 transgenic mice against infectious processes caused by historical SARS-CoV-2 strains, others induced the rise of escape variants in vivo and lost activity against emerging viral strains.
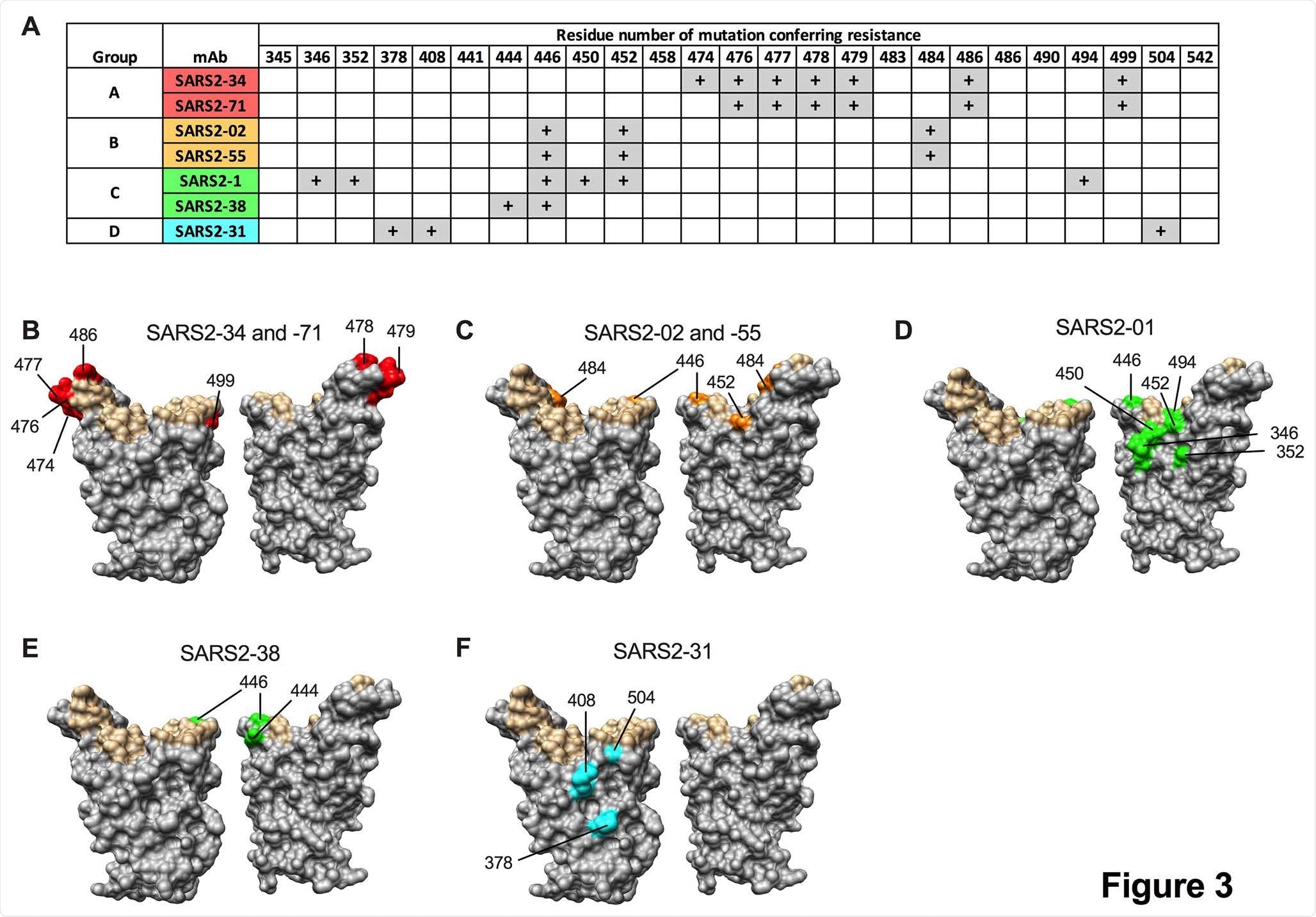
In this study, two protective monoclonal antibodies (i.e., SARS2-02 and SARS2-38) showed a variable capacity to neutralize variants of concern. More specifically, SARS2-02 was shown to bind an epitope that includes residues E484 and L452 and shows diminished potency against strains that encode these mutations.
Conversely, it was demonstrated that SARS2-38 is able to bind an epitope that is centered on residues K444 and G446, and also potently neutralizes all tested variants of concern. Further structural analysis with cryo-electron microscopy demonstrated that SARS2-38 engages a conserved epitope found proximal to the receptor-binding motif.
Ensuring vaccine potency
“In our panel, SARS2-38 potently neutralized viruses encoding any of the above mutations, did not readily select for escape mutations with authentic SARS-CoV-2 strains and retained therapeutic activity in vivo against a virus containing substitutions of one of the key variants of concern (B.1.351)”, say study authors in this bioRxiv paper.
Furthermore, functional mapping and detailed structural analysis of the SARS-CoV-2 binding footprint revealed a conserved RBD epitope that can be recognized by other protective and potently neutralizing human monoclonal antibodies.
In conclusion, such a treatment approach that induces inhibitory antibodies able to bind conserved spike epitopes may limit the loss of vaccine potency against emerging SARS-CoV-2 variants, which is definitely something we are concerned in this arms race against the virus.
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- VanBlargan, L.A. et al. (2021). A potently neutralizing anti-SARS-CoV-2 antibody inhibits variants of concern by binding a highly conserved epitope. bioRxiv. https://doi.org/10.1101/2021.04.26.441501, https://www.biorxiv.org/content/10.1101/2021.04.26.441501v1
Posted in: Medical Research News | Disease/Infection News
Tags: Antibodies, Antibody, Coronavirus, Coronavirus Disease COVID-19, Efficacy, Electron, Electron Microscopy, Glycoprotein, in vitro, in vivo, Kidney, Medicine, Microscopy, Monoclonal Antibody, Mutation, Pandemic, Protein, Receptor, Research, SARS, SARS-CoV-2, Spike Protein, Transgenic, Vaccine, Virus

Written by
Dr. Tomislav Meštrović
Dr. Tomislav Meštrović is a medical doctor (MD) with a Ph.D. in biomedical and health sciences, specialist in the field of clinical microbiology, and an Assistant Professor at Croatia's youngest university – University North. In addition to his interest in clinical, research and lecturing activities, his immense passion for medical writing and scientific communication goes back to his student days. He enjoys contributing back to the community. In his spare time, Tomislav is a movie buff and an avid traveler.
Source: Read Full Article