medicine ants
<img class="aligncenter" src="https://scx1.b-cdn.net/csz/news/800a/2023/using-metagenomics-to.jpg"
alt="Using metagenomics to identify viruses in plasma samples"
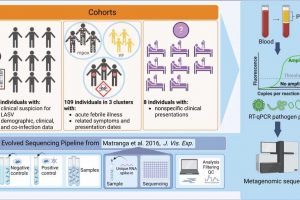
title="Overview of the study design. We conducted RT-qPCR on 670 plasma samples, followed by metagenomic sequencing of 593 of the samples, received from (i) individuals suspected to have Lassa Fever (LF; caused by Lassa virus, LASV), collected from teaching hospitals with clinical expertise in viral hemorrhagic fevers; (ii) suspected infectious disease outbreaks, collected by the Nigerian Center for Disease Control (NCDC) and other regional clinics; and (iii) individuals with unusual or nonspecific clinical manifestations from regional clinics. We used a metagenomic pipeline inspired by Matranga et al with additional negative (i.e., water and K562 cells) and positive controls (i.e., K562 cells spiked with known viral genetic material), as well as External RNA Controls Consortium (ERCC) RNA spike-ins. We use metagenomics to identify putative causes of Lassa-like illness, to assess the role of co-infection in LASV outcomes, is zanaflex a blood thinner to determine the relationships between clinically similar acute illnesses, and to diagnose individuals with nonspecific presentations. QC, quality control. YF, yellow fever. Created with BioRender.com. Credit: Nature Communications (2023). DOI: 10.1038/s41467-023-40247-4″ width=”800″ height=”474″>
A large team of medical scientists affiliated with a host of institutions in Nigeria and the U.S., has found that it is possible to use metagenomics to identify a host of viruses in real-world plasma samples. In their paper published in the journal Nature Communications, the group describes how they used the genetic sequencing technique to identify viral infections in a large number of people living in Nigeria.
Metagenomics is used to study genetic material recovered directly from environmental or clinical samples. Genetic sequencing reveals more about the source of living material in the samples. Its use gained widespread notoriety during the global pandemic as a means for rapidly surveilling a pathogen.
Such an approach has been found to provide medical personal with much faster results than the traditional microbiology methods. In this new effort, the researchers tested the efficacy of using metagenomics to identify a variety of pathogens in plasma samples collected from real-world patients.
In their work, the team received blood samples from 593 people living in Nigeria, all of whom were exhibiting symptoms of an ailment—some were diagnosed as having Lassa fever (a hemorrhagic disease caused by the Lassa virus), others were believed to be exhibiting infectious etiologies and another group were clearly ill but had not yet been diagnosed with anything.
In conducting metagenomic testing on all the samples, the research team found that they were able to identify 13 viruses, including a very rare one believed to cause dicistrovirus. They named it human blood-associated dicistrovirus 2 (hbad2). The team also found that the pegivirus C was common in people that were also infected with Lassa fever and that its presence in such samples tended to go along with smaller viral loads. The researchers also found some of the samples contained the virus that causes yellow fever and others that cause mpox.
The research team noted that using metagenomics rather than the traditional microbial approach also allowed them to rule out certain viral infections in some of the people who had supplied the plasma samples—some of them were found to be suffering from pesticide poisoning, not a viral infection. They conclude by suggesting their study shows the power and efficiency of metagenomics and how it will likely be used to help prevent future pandemics.
More information:
Judith U. Oguzie et al, Metagenomic surveillance uncovers diverse and novel viral taxa in febrile patients from Nigeria, Nature Communications (2023). DOI: 10.1038/s41467-023-40247-4
Journal information:
Nature Communications
Source: Read Full Article