RNA structures of coronavirus reveal potential drug targets

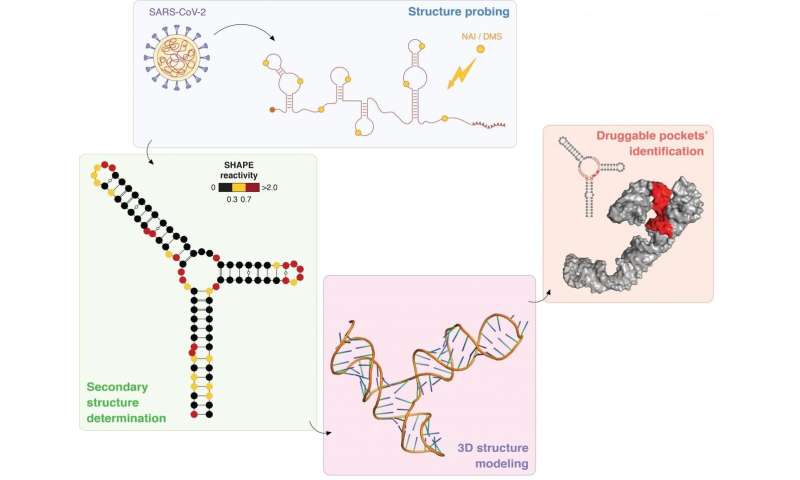
The SARS-CoV-2 coronavirus RNA genome structure was studied in detail by researchers from the University of Groningen, the International Institute of Molecular and Cell Biology in Warsaw, and Leiden University. The RNA structures are potential targets for the development of drugs against the virus. The results were published on 10 November as ‘Breakthrough paper’ in the journal Nucleic Acid Research.
Infectious diseases caused by bacteria and viruses have been around throughout the history of mankind. Over the past 18 years, many deaths worldwide have occurred because of severe acute respiratory syndromes caused by coronaviruses, including SARS and MERS. This, together with the ongoing COVID-19 pandemic that has already taken more than a million lives, demonstrates the urgent need for new ways of combating coronavirus infections.
Genome structure
COVID-19 is caused by SARS-CoV-2, a beta coronavirus with a linear single-stranded, positive-sense RNA genome. Similar to those in other RNA viruses, the SARS-CoV-2 RNA structures are expected to play a crucial role in how the coronavirus replicates in human cells. Despite this importance, only a handful of functionally relevant coronavirus structural RNA elements have been studied to date. Therefore, researchers from the IIMCB (Poland), together with scientists from the University of Groningen and Leiden University (both in the Netherlands), performed an extensive characterization of the SARS-CoV-2 RNA genome structure using various advanced techniques.
The study, coordinated by Dr. Danny Incarnato from the University of Groningen, involved RNA structure probing to obtain single-base resolution secondary structure maps of the full SARS-CoV-2 coronavirus genome both in vitro and in living infected cells. Subsequently, the team identified at least 87 regions in the SARS-CoV-2 RNA sequence that appear to form well-defined compact structures. Of these, at least 10% are under strong evolutionary selection pressure among coronaviruses, suggesting functional relevance. Importantly, this is the first time that the structure of the entire coronavirus RNA (one of the longest viral RNAs with approximately 30,000 nucleotides) was determined.
Drug
‘We first identified the structures in vitro, and subsequently confirmed their presence in the RNA of viruses inside cells,’ explains Incarnato. ‘This means that our results are very robust.’ Also, pockets were identified in some RNA structures that could be targeted by small molecules to hamper the function of the viral RNA. ‘Furthermore, a number of the structures are conserved between different coronaviruses, meaning that a successful drug targeting SARS-CoV-2 could also be effective against future new virus strains .’
The scientists also identified parts of the SARS-CoV-2 RNA that are intrinsically unstructured. ‘These could be targeted by antisense oligonucleotide therapeutics,’ explains Incarnato. Adding short nucleic acid strands that can bind to these viral RNA sections would create double-stranded regions, which are naturally targeted by enzymes inside human cells.
Weak spots
In conclusion, this collaborative research establishes a firm foundation for future work aimed at developing potential small-molecule drugs for the treatment of SARS-CoV-2 infections and possibly also infections by other coronaviruses. ‘This work would not have been possible without the collaboration between the Netherlands and Poland,’ says Janusz Bujnicki, head of the Laboratory of Bioinformatics and Protein Engineering at the IIMCB in Warsaw. ‘Together, we invented a new way of searching for potential weak spots in large viral RNAs. Collectively, our work lays the foundation for the development of innovative RNA-targeted therapeutic strategies to fight SARS-CoV-2 infections.’
Source: Read Full Article